
在撰写有关异常检测器开发的文章时,我实现了一种称为增量生长神经气体的算法。
在 苏联文学 在Internet的俄罗斯部分,该主题的覆盖面很差,只有一篇文章 ,甚至在此算法的应用之后。
那么增量增长的神经气体算法到底是什么呢?
引言
IGNG与GNG一样,是一种自适应聚类算法。
Prudent和Ennadji在2005年的一篇文章中介绍了该算法本身。
像GNG一样,有很多数据向量 或产生功能 ,它从随机分布的数据(参数 -时间或样本中的样本编号)。
该算法不对这些数据施加其他限制。
但是内部与GNG有很大不同。
该算法也很有趣,因为它比GNG模型的神经发生精确得多。
算法说明
该算法将大量数据分解为群集。
与GNG相比,其优点是收敛速度更高。
该算法所基于的思想:
- 自适应共振理论 :首先,搜索最近的神经元,如果差异不超过阈值(“警戒参数”),则调整权重,否则,将更改数据空间中的神经元坐标。 如果未克服阈值,则会创建新的神经元,以更好地近似数据样本的值。
- 连接和神经元都具有年龄参数(GNG仅具有连接),该参数起初为零,但随着您的学习而增加。
- 神经元不会立即出现:首先出现胚胎(或生发神经元),其年龄随着每次迭代而增加,直到成熟。 训练后, 只有成熟的神经元才参与分类 。
主循环
工作始于空图。 参量 由训练样本的标准偏差初始化:
其中: -样本坐标之间的平均值。
每个步骤的主循环都会减小该值 ,它是接近度阈值,用于计算先前的聚类质量级别和通过IGNG程序进行聚类后获得的级别之间的差异。
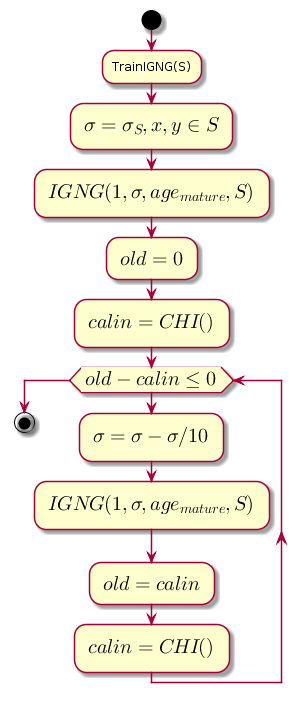
图表代码。@startuml start :TrainIGNG(S); :<latex>\sigma = \sigma_S,x,y \in S</latex>; :<latex>IGNG(1, \sigma, age_{mature}, S)</latex>; :<latex>old = 0</latex>; :<latex>calin = CHI()</latex>; while (<latex>old - calin \leq 0</latex>) :<latex>\sigma=\sigma - \sigma / 10</latex>; :<latex>IGNG(1, \sigma, age_{mature}, S)</latex>; :<latex>old = calin</latex>; :<latex>calin = CHI()</latex>; endwhile stop @enduml
CHI是Kalinsky-Kharabaz指数,显示了聚类的质量:
其中:
- -数据样本数。
- -簇数(在这种情况下为神经元数)。
- -内部弥散矩阵(神经元坐标之间的平方距离和所有数据的平均值之和)。
- -外部色散矩阵(所有数据和最近的神经元之间的距离的平方和)。
索引值越大越好,因为如果聚类之后和索引之前的索引之间的差为负,则可以假定索引变为正并超过了前一个索引,即 群集成功完成。
IGNG程序
这是算法的基本过程。
它分为三个互斥的阶段:
- 找不到神经元。
- 发现了一个令人满意的神经元。
- 找到了两个满足神经元条件的东西。
如果条件之一通过,则不执行其他步骤。
首先,在神经元中搜索最佳近似数据样本:
在这里 -距离计算函数,通常是欧几里德度量 。
如果找不到神经元,或者距离数据太远,即 不符合邻近标准 ,将创建一个新的胚胎神经元,其坐标等于数据空间中样本的坐标。

如果通过了邻近性检查,则以相同的方式搜索第二个神经元,并检查与数据样本的邻近性。
如果未找到第二个神经元,则会创建它。

如果发现两个神经元满足与数据样本接近的条件,则根据以下公式校正其坐标:
其中:
- -适应步骤。
- 是神经元的数量。
- -神经元邻域功能 与优胜者神经元(在这种情况下,对于直接邻居,它将返回1,否则将返回0,因为适应步骤用于计算 仅对于直接邻居为非零)。
换句话说,获胜神经元的坐标(权重)更改为 ,以及它的所有直接邻居(通过图形的一条边与之相连的邻居)在 在哪里 -更改前相应神经元的坐标。
然后,在两个获胜的神经元之间创建连接,如果已经创建,则重置其年龄。
所有其他关系的年龄正在增加。
所有年龄超过常数的通讯 被删除。
之后,将所有孤立的(与他人无关的) 成熟神经元移除。
与获胜神经元相邻的直接神经元的年龄正在增加。
如果任何种系神经元的年龄超过 他成为一个成熟的神经元。
最终图形仅包含成熟的神经元。
可以将完成以下IGNG程序的条件视为固定数量的循环。
该算法如下图所示(图片可点击):
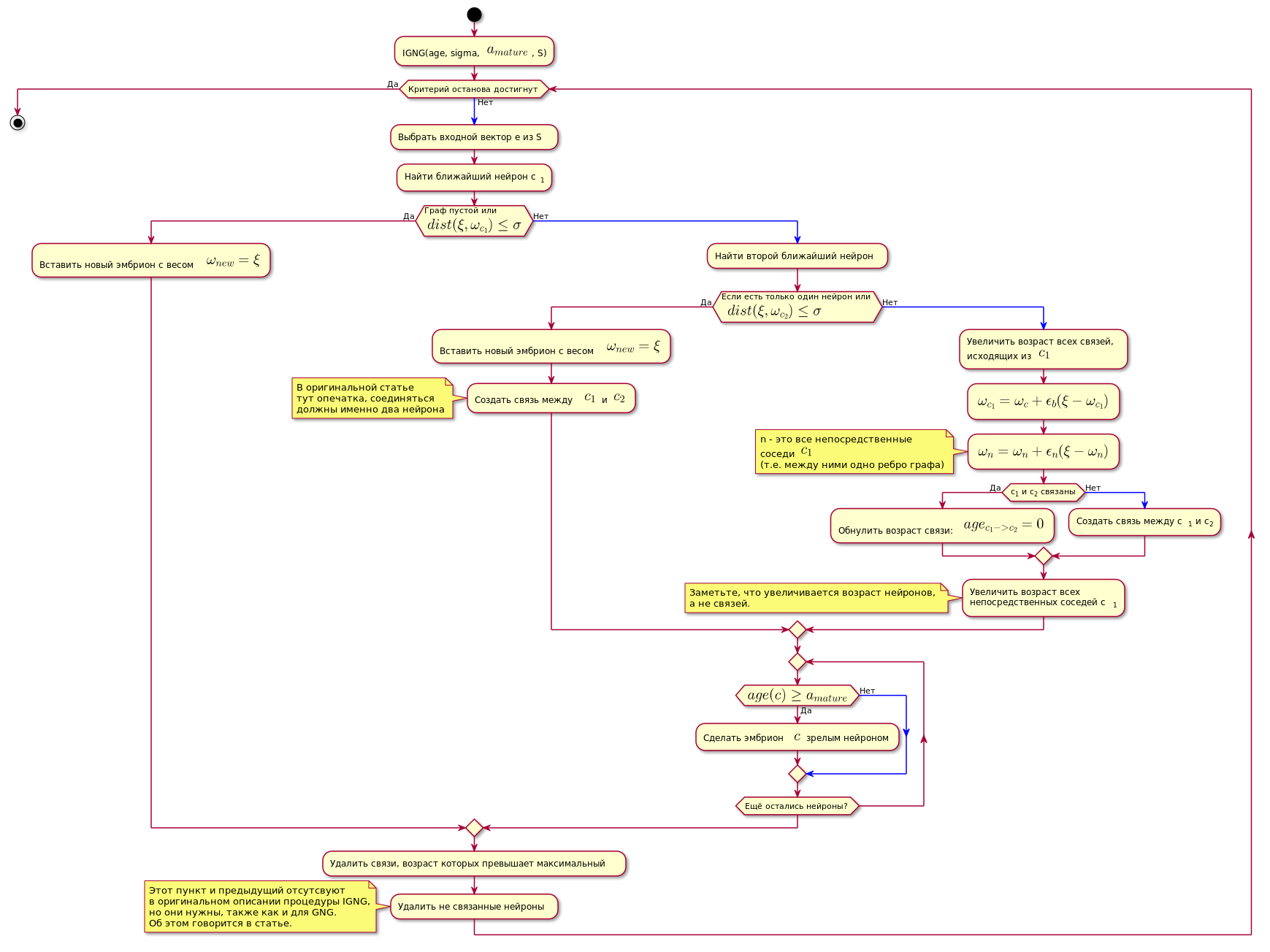
图表代码。 @startuml skinparam nodesep 10 skinparam ranksep 20 start :IGNG(age, sigma, <latex>a_{mature}</latex>, S); while ( ) is () -[#blue]-> : e S; : c<sub>1</sub>; if ( \n<latex>dist(\xi, \omega_{c_1}) \leq \sigma</latex>) then () : <latex>\omega_{new} = \xi</latex>; else () -[#blue]-> : ; if ( \n <latex>dist(\xi, \omega_{c_2}) \leq \sigma</latex>) then () : <latex>\omega_{new} = \xi</latex>; : <latex>c_1</latex> <latex>c_2</latex>; note , end note else () -[#blue]-> : ,\n <latex>c_1</latex>; :<latex>\omega_{c_1} = \omega_c + \epsilon_b(\xi - \omega_{c_1})</latex>; :<latex>\omega_n = \omega_n + \epsilon_n(\xi - \omega_n)</latex>; note n - <latex>c_1</latex> (.. ) end note if (c<sub>1</sub> c<sub>2</sub> ) then () : : <latex>age_{c_1 -> c_2} = 0</latex>; else () -[#blue]-> : c<sub>1</sub> c<sub>2</sub>; endif : \n c<sub>1</sub>; note , , . end note endif repeat if (<latex>age(c) \geq a_{mature}</latex>) then () : $$ ; else () -[#blue]-> endif repeat while ( ?) endif : , ; : ; note IGNG, , GNG. . endnote endwhile () stop @enduml
实作
该网络是使用NetworkX图形库以Python实现的。 下面给出了从上一篇文章的原型中删除代码的信息。 对于代码也有简短的解释。
如果有人对完整代码感兴趣,那么这里是指向存储库的链接 。
算法示例:

大部分代码 class NeuralGas(): __metaclass__ = ABCMeta def __init__(self, data, surface_graph=None, output_images_dir='images'): self._graph = nx.Graph() self._data = data self._surface_graph = surface_graph